Artificial Enzymes
Hydrogen production catalyzed by bio-inspired catalysts is still associated with high overvoltages so that optimization need to be achieved. For that, it is crucial to better understand how the interactions with peptidic residues stabilize and beyond control the reactivity and catalytic activity of soft organometallic centers in aqueous solutions. In this perspective, a bio-synthetic approach, i.e using peptides or proteins as coordinates instead of synthetic ligands, is very attractive but remains to be investigated regarding the design of artificial hydrogenases.
Artificial metalloenzymes result from the association of a catalytically active synthetic organometallic or coordination compound with a biomacromolecular host, protein or DNA. A priori a minimum of three effects contribute to the generally superior catalytic properties observed for (inorganic) catalytic co-factors when embedded inside a protein: i) site isolation, preventing aggregation; ii) substrate control - channels regulate the amount of substrate present around the catalyst, as well as protect it from unwanted substrates; iii) specific protein-complex interactions resulting in the stabilization of a specific geometry of the complex with enhanced catalytic properties (entatic states), via e.g. hydrogen bonding.
In a first study, in collaboration with Christine Cavazza (IBS), we have investigated the reactivity of a variety of water-soluble iron, ruthenium and manganese carbonyl complexes towards lysozyme. This protein was selected on the basis of its reported ability to incorporate a {Ru(p-cymene)}2+ center. In most cases however, binding to lysozyme was not specific and no defined adduct could be isolated but we could purify and characterize, at the spectroscopic and structural level, a {Mn(CO)3}+ derivative of lysozyme. Furthermore, we have found that this compound is able to transfer the {Mn(CO)3}+ group to a nickel complex, thus forming a structural mimic of [NiFe] hydrogenases active site similar to the functional nickel-ruthenium electrocatalyst described above.
Unfortunately, the dinuclear compound, once formed, escape from the protein scaffold.

Figure 1: Structure of the covalent adduct [lysozyme-{MnI(CO)3}] and recativity with the nickel complex [Ni(xbsms)].
In order to accommodate a dinuclear cluster within a protein framework and thus design a true artificial enzyme, we used a second, even more biomimetic, approach. Its biological relevance resides in all three components of the system, i.e. the choice of synthetic co-factor and scaffold protein as well as the binding mode between the two. First, the synthetic catalyst, featuring two cyanide ligands, ([(µ-S(CH2-X-CH2)S)Fe2(CO)4(CN)2]2-) (X = CH2, O or NH) is a close mimic of the active centre of the [FeFe]-hydrogenase and it is used in such an application for the first time. Second, HydF was used as the protein host candidate as it has been established as a key component of the hydrogenase maturation machinery: it functions as a scaffold protein involved in the assembly of the diiron centre and its transfer to apo [FeFe]-hydrogenase, for final maturation of the latter. It is namely anticipated that HydF contains a well-designed cavity for binding an Fe2(CO)x(CN)y complex and indeed, HydF is able to bind one equivalent of the above mentioned diiron clusters. Based on a complete set of spectroscopic (UV-visible, FTIR, EPR) spectroscopies and related DFT computations, run in collaboration with S. Gambarelli and J.-M. Mouesca from INAC (CEA/Université Joseph Fourier) and colleagues from the Lubitz group at the Max-Plank Institute for Chemical energy Conversion (Mülheim, Germany), we could establish that the dinuclear cluster binds via a bridging cyanido ligand to the [4Fe-4S] cluster of HydF in which only three Fe atoms are ligated by a cysteine residue while the fourth one displays an accessible coordination site. This latter site allowed for direct association between the dinuclear synthetic complex and the [4Fe-4S] cluster.
More importantly, based on strong similarities observed in the FTIR spectra recorded for this artificially loaded HydF protein as compared to natural HydF, expressed in the presence of the other maturation enzymes, it was argued that this artificial enzyme, with a bridging cyanido ligand, was a representative model also for the natural form of HydF primed for activation of apo-[FeFe] hydrogenase. Indeed the artificially loaded HydF was found capable of transferring its synthetic co-factor to the apo-form of the [FeFe] hydrogenase enzyme. Full activation of [FeFe]-hydrogenase was achieved exclusively using the HydF hybrid protein containing the mimic with an azadithiolate bridge, confirming the presence of this ligand in the active site of native [FeFe]-hydrogenase.
MPI-CEC, Mülheim, Germany
Figure 2 : Biomimetic assembly mechanism bypassing much of the biosynthesis of the [FeFe]-hydrogenase enzyme, resulting in the introduction of synthetic analogues of the enzyme's [2Fe] subsite and its activatation when X = NH. Reproduced from “Bioinorganic chemistry: Enzymes activated by synthetic components” Ryan D. Bethel & Marcetta Y. Darensbourg, Nature 2013 499, 40–41
Cobaloxime-Based Artificial Hydrogenases
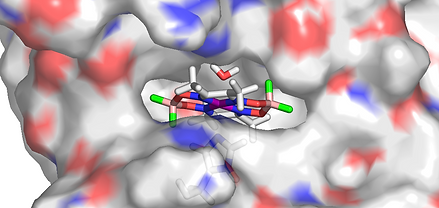
Sperm-whale myoglobin (SwMb) has been used as a host protein for the construction of a variety of artificial enzymes that catalyze oxidation of organic substrates. Schiff-base complexes of chromium(III), manganese(III), iron(III) and copper(II) have been inserted within the cavity of the apo-SwMb host where they bind to a histidine residue. In most cases, this is His93, which is the natural ligand of hemin in holo-SwMb, and which coordinates the square planar Schiff-base complexes in the axial position, as revealed by X-ray crystallography. Surprisingly, this strategy has never been used to prepare artificial hydrogenases. In our group, we develop cobaloximes. These efficient H2-evolving catalysts possess square-planar geometry similar to Schiff-base complexes together with good affinity in their axial position for N-heterocyclic bases. We could show that cobaloximes moieties such as {Co(dmgH)2} or {Co(dmgBF2)2} can be easily installed in myoglobin through coordination to only one of the two histidines available in the binding pocket.
The resulting biohybrids contain one cobalt complex per polypeptidic chain and all spectroscopic data indicate that they exist as pure molecularly defined species. EPR spectroscopy proves coordination of a histidine residue in axial position of the cobalt complex and CD spectroscopy indicates that the uptake of these synthetic cofactors results in an increase in the structuring of a-helices as observed during the maturation of holo-SwMb. The reaction of the SwMb.cobaloxime biohybrids with one equivalent of hemin at room temperature does not result in the replacement of the cobalt complex by hemin, indicating that the formation of the SwMb.cobaloxime biohybrids are irreversible processes or occur with a very high association constant. All these data support coordination of the cobaloximes onto His93 and their location within the SwMb cavity. QC/MM docking calculations confirmed the possible location of the cobaloxime moiety within the protein pocket and indicated coordination preference of His93 over the other histidine residue (His64) present in the vicinity. Structural data extracted from these calculations fit well with those obtained from EXAFS measurements on both biohybrids. In both cases, the cobalt center appears to be hexacoordinated with a water molecule filling the coordination sphere of the cobaloxime complexes bound to the protein via axial His93. The hydrophobic environment of the cobaloxime when inserted within the myoglobin cavity is supported by the comparison of the g-values extracted from EPR spectra of one biohybrid with those already reported for the {Co(dmgBF2)2} moiety in various compounds or media. Electrochemical measurements on biohybrid-modified CNT electrodes demonstrated redox activity of the cobalt centers in both biohybrids. The potential of the Co(II)/Co(I) couple is significantly affected by the protein environment with a 100 mV negative shift as compared to the free cobaloxime moieties under similar conditions. Solution assays based either on thermal reduction of the catalyst by Eu(II) complexes or light-driven reduction of the catalyst by photogenerated DAFH radical or [Ru(bipy)3]*2+ have been used to assess the catalytic properties of the biohybrids. Both biohybrids possess catalytic activity for H2 evolution in aqueous pH 7 solutions and compare well under such conditions with the other systems reported in the literature, all based on dithiolate-bridged hexacarbonyl diiron catalytic units covalently attached to polypeptides or incorporated within cytochrome c. Importantly these cobaloxime-based artificial hydrogenases display catalytic activity for H2 evolution with low overpotential requirement (~200 mV), which is key for further biotechnological application since this parameter directly impacts the energetic efficiency of electrochemical devices. The operational stability, another crucial requirement for technological implementation, appears limited to few turnovers, as observed for most artificial hydrogenases described so far. This can however be improved in the future through enhanced electron communication with the electron donor observed with H2-evolving catalysts immobilized onto electrode materials or through active site engineering.





